The following email was sent to all members of project ua6 on 18/8/2017 by Clare Richards.
Dear ua6 member,
As you are hopefully aware, ua6 is currently scheduled for migration to NCI’s new /g/data1 filesystem on Tuesday, 22 August. You will not be able to access data while the move is in progress.
The schedule and information about the migration process can be found here https://opus.nci.org.au/display/NDM/NCI+Data+Migration (use your NCI login).
In the lead-up to migration, the ua6 data managers have identified a number of files which were duplicated during CMIP5 data download. Paola Petrelli of the Climate CoE CMS team has compiled a list of directories which are identical copies on the system, that is, they contain files that have the same tracking ID but are a superseded version, or even exactly the same files downloaded from different ESGF servers. We plan to remove these duplicates at the same time as the migration outage.
To see the list of ~9,000 identified directories, please visit this page: https://opus.nci.org.au/display/CMIP/CMIP5+replica+clean+up
The logic behind duplicate identification, and the final list of candidates for deletion, has been reviewed separately by data managers from the Bureau of Meteorology and CSIRO, and NCI have approved the data removal.
What do you need to do?
- If you are currently using the ARCCSSive database to access CMIP5 data, there will be no impact on you, the database has already been updated to point to the “to keep” versions of each file.
- If you use the DRSv2 structure, please be advised that the DRS tree will need to be rebuilt following the deletion. In the interim it will be moved to “DRSv2_legacy” and a new DRSv2 tree built from scratch. We will notify you when the new DRS tree structure is ready in the days following clean up and migration.
- If you access data directly on the filesystem using absolute paths, we recommend you review the list of deletion candidates attached to the above wiki page
We anticipate these changes will have minimal impact on the community but please provide any feedback or concerns by 5pm on Monday 21/8/2017 to Clare.Richards@anu.edu.au and Paola.Petrelli@utas.edu.au.
Please note that ua6 access points in THREDDS and the VDI (CWSLab), will be unavailable from the evening of August 21st. Services will return overnight the day migration and clean up is completed, i.e. 24-48 hours later. This includes ua6_4 NRM data.
Thank you for your patience during this process. We will notify you when the data is available again.
Kind regards,
The ua6 data managers
Scott's presentation from today is at https://docs.google.com/presentation/d/1hKx1cdTF5WlgGLxRYErMfsGN4wKii_rXVBeOl4AtvKI/edit?usp=sharing
Documentation for the postgres branch of ARCCSSive is at http://arccssive.readthedocs.io/en/postgres/
Install instructions and an overview of the SQLalchemy model classes is at http://arccssive.readthedocs.io/en/postgres/v2.html
The annual ESGF user survey has been completed for 2016 and the results compiled. A document summarising the results is attached here.
The agenda and dial-in details for the ESGF F2F meeting in Washington DC, Dec 6-9 2016 have been released.
Please note the timezone is GMT-5 (=UTC-5).
Agenda
Current version of the agenda (23/11/2016) is attached.
Also find attached the list of abstracts.
Call in details
REMOTE PARTICIPANTS
GUIDANCE for EMAIL REGISTRATION
For anyone participating in the ESGF F2F conference via REMOTE LOGIN (WebEx), please follow this guidance:
· After login to the meeting via WebEx (see link and access code below)
· Please email Angela Jefferson (jefferson9@llnl.gov) with the following details:
o First and Last Name
o Company Affiliation
o Email address
2016 ESGF F2F - Marriott Metro Center, Washington, DC
Every Monday, Tuesday, Wednesday, Thursday, Friday, from Tuesday, December 6, 2016, to Friday, December 9, 2016
7:30 am | Eastern Standard Time (New York, GMT-05:00) | 11 hrs
Meeting number (access code): 801 978 147
Meeting password: esgf
When it's time, join the meeting.
The following email has been circulated to "cmip-users@llnl.gov".
In the future you might expect to receive similar information perhaps a couple of times a year. If you do not want to receive emails of this kind, please let us know so that we can remove you from the mailing list.
I have installed and played around with pyesgf http://esgf-pyclient.readthedocs.org/en/latest/index.html a python module that allows to interact with the ESGF API using python.
Basically you can build a search passing on constraints as you can do online and get back the search results.
The advantage of using this is that you can decide how you want these results to be retrieved, for example currently Lawson scripts and also my own python scripts where retrieving a wget file which then was parsed to extract the information on available files such as checksum, download url etc.
With pyesgf you have different options to retrieve the search results:
wget
opendap aggregation
a "list of datasets" object DatasetResult and each of these has a "list of files" object FileResult
Following are some of the properties that can be retrieve for each of these
DatasetResult:
methods: 'context', 'dataset_id', 'download_url', 'file_context', 'index_node', 'json', 'las_url', 'number_of_files', 'opendap_url', 'urls'
Where all the info is hidden is the "json" attribute (a python dictionary), that's the actually entire response from the server, so you get from it much more information including the version. The dict keys are:
[u'index_node', u'version', u'dataset_id_template_', u'cf_standard_name', u'number_of_aggregations', u'south_degrees', u'drs_id', u'cmor_table', u'replica', u'west_degrees', u'master_id', u'height_top', u'id', u'datetime_stop', u'height_bottom', u'access', u'realm', u'data_node', u'title', u'description', u'instance_id', u'height_units', u'variable_long_name', u'metadata_format', u'variable_units', u'score', u'datetime_start', u'number_of_files', u'_version_', u'size', u'type', u'ensemble', u'product', u'experiment', u'format', u'timestamp', u'time_frequency', u'_timestamp', u'variable', u'east_degrees', u'experiment_family', u'forcing', u'institute', u'north_degrees', u'project', u'url', u'model', u'latest']
FileResult:
methods: 'checksum', 'checksum_type', 'context', 'download_url', 'file_id', 'filename', 'index_node', 'json', 'las_url', 'opendap_url', 'size', 'tracking_id', 'urls'
json dictionary keys:
[u'index_node', u'version', u'dataset_id_template_', u'cf_standard_name', u'cmor_table', u'replica', u'master_id', u'dataset_id', u'id', u'size', u'instance_id', u'realm', u'data_node', u'title', u'description', u'drs_id', u'variable_long_name', u'metadata_format', u'variable_units', u'score', u'_version_', u'forcing', u'type', u'ensemble', u'product', u'experiment', u'format', u'timestamp', u'time_frequency', u'variable', u'_timestamp', u'institute', u'checksum', u'experiment_family', u'project', u'url', u'tracking_id', u'checksum_type', u'model', u'latest']
ESGF search records contain various ids. These include:
- id: internal identifier so that SOLr can keep track of each record. Currently the instance_id + index node
- drs_id: DRS identifier without the version number. Effectively an identifier for each dataset (including all versions and replicas)
- master_id: The same as drs_id. I think this was created because of confusion about the purpose of DRS and drs_id. E.g. it's not clear whether a DRS identifier includes a version
- instance_id: .. I.e. the DRS including the version number
The dataset_id listed above is the first, but we could potentially retrieve any of them, Examples are
id - cmip5.output1.MIROC.MIROC5.historical.mon.atmos.Amon.r5i1p1.v20120710|aims3.llnl.gov which is the dataset_id property
drs_id - cmip5.output1.MIROC.MIROC5.historical.mon.atmos.Amon.r5i1p1
master_id - cmip5.output1.MIROC.MIROC5.historical.mon.atmos.Amon.r5i1p1
instance_id - cmip5.output1.MIROC.MIROC5.historical.mon.atmos.Amon.r5i1p1.v20120710
This is what i've managed up to now:
I've listed all the netcdf/cmip files under
/g/data2/ua6_jxa900/DKRZ/
and
/g/data2/ua6_jxa900/LLNL
simply using this script
I'm doing the same currently for the BADC directory but it's waiting in the queue
Then I've used my old search_CMIP5_replica.py script to extract only the ensembles, that reduced the lines to something manageable.
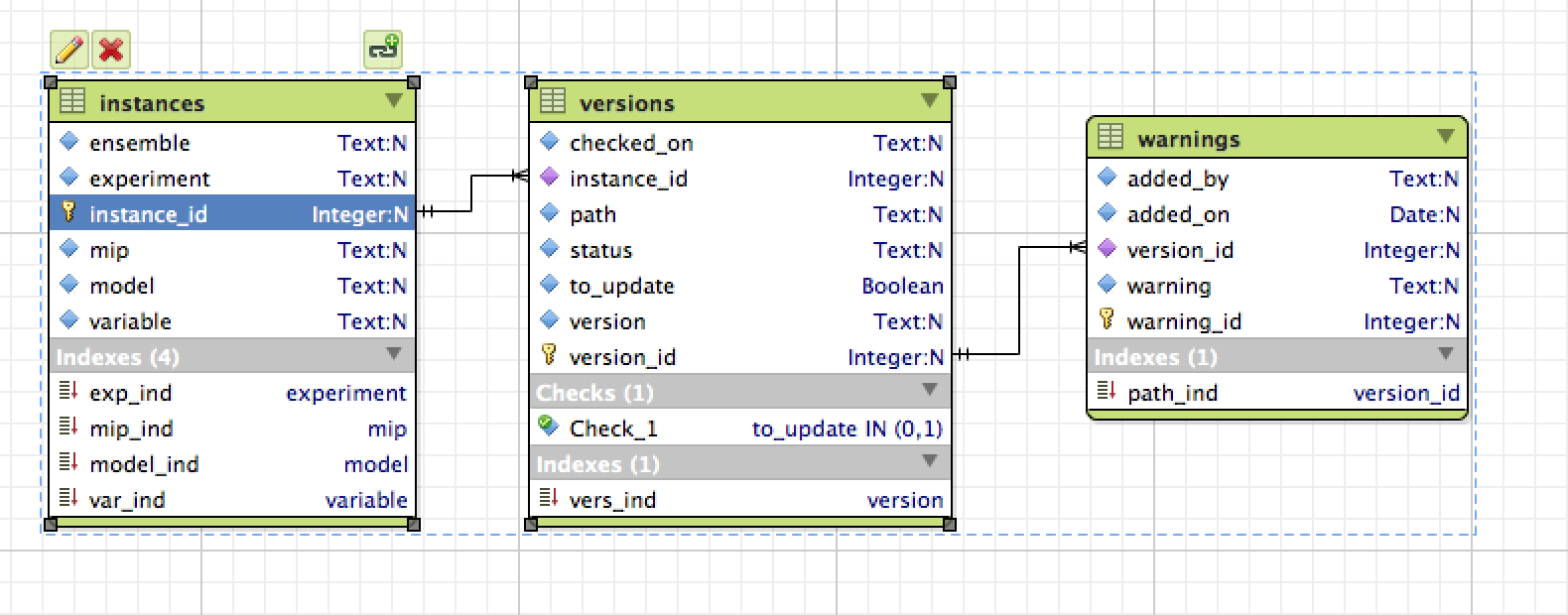
Finally I'm creating a sqlite database to list them, the schema is
This is the same schema I'm using for the data in unofficial, I've got python script to update the version information and to add user warnings.
I will add them to my CMIP-utils repo, but probably on Monday once I checked them properly.
As "latest" I mean not the latest on raijin, but the latest published, which can't be determined yet until the data is back online, the date checked_on refers to when a "latest/noreplica" search on ESGF was last done and the files checksums compared to what available on raijin.
Both databases are in
/g/data1/ua6/unofficial-ESG-replica/tmp/tree/
bulk_test.db
new_CMIP5_test.db
Unfortunately, I forgot I was cutting a bit of the path that was working fine when everything was coming from unofficial and it's an issue instead for the bulk data, so the current database doesn't have the directory roots, I'm re-running it now. It's relatively fast, but it'll be waiting int he queue for a while. Also the versions at elast for LLNL data don't mean anything out of a local context. So something still needs to be done to compare them to what we have already to determine if they are more or less recent, but also their version number.
I'll be looking into this later working only on subsets I'm interested into, and dd the info to the database. But you might have a better more systematic way to solve this.
Scott as create a python module using SQLalchemy to extract the info ARCCSSive this is a work in progress, in fact I've just added some functionality to check warnings, but hasn't been merged yet. Any feedback appreciated.
I've tried to download some data from pcmdi9 using the scripts created by Lawson. The data transfer went ok, but the md5 checksum fails. Turns out that they are using a different hashing algorithm SHA256, but when executing the wget file at the end of the download still calculates MD5. So the hash ends up being different from the one listed in the same wget script. The original wget script has the correct code which check if the node has passed an md5 or sha256 hash, but Lawson scripts insert a section that calculates md5sum by default.
To be more clear, this is what a new file line looks like in a wget script
'tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc' 'http://aims3.llnl.gov/thredds/fileServer/cmip5_css01_data/cmip5/output1/LASG-CESS/FGOALS-g2/rcp85/3hr/atmos/3hr/r1i1p1/v2/tas/tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc' 'SHA256' '8f67c9f75395212495cbe9138863ba77012f6749457dbed82542b06525391e13'
this what I’m getting by running wget for one file
tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc ...Already downloaded and verified
done
ERROR file 'tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc' MD5 checksum test failed
This is from the modified "trailer-part" of the wget script after the download is completed:
if [ "x8f67c9f75395212495cbe9138863ba77012f6749457dbed82542b06525391e13" != "x" ]
then
newMd5=`md5sum tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc | awk '{print $1}' -`
if [ "${newMd5}" != "8f67c9f75395212495cbe9138863ba77012f6749457dbed82542b06525391e13" ]
then
echo "ERROR file 'tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc' MD5 checksum test failed" >> /home/581/pxp581/cm5-download-errors.log
echo "ERROR file 'tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc' MD5 checksum test failed" 1>&2
exit 99
fi
fi
Changing cm5-download-mod2.sh so that it adds this bit of code after "then" seems to fix it
case $chksum_type in
md5)
newMd5=`md5sum tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc | cut -d ' ' -f1 `;;
sha256)
newMd5=`sha256sum tas_3hr_FGOALS-g2_rcp85_r1i1p1_203901010300-204001010000.nc |cut -d ' ' -f1`;;
esac
So around line 690 of cm5-download-mod2.sh
echo " if [ \"x${md5}\" != \"x\" ]" >> $oFile
echo " then" >> $oFile
echo " case \$chksum_type in" >> $oFile
echo " md5)" >> $oFile
echo " newMd5=\`md5sum ${lfn} | cut -d ' ' -f1\`;;" >> $oFile
echo " sha256)" >> $oFile
echo " newMd5=\`sha256sum ${lfn} | cut -d ' ' -f1\`;;" >> $oFile
echo " esac" >> $oFile
echo " " >> $oFile
echo " if [ \"\${newMd5}\" != \"${md5}\" ]" >> $oFile
echo " then" >> $oFile
.....
Changes in servers' names
My version of the bash scripts now works again fine and can download files, the problem is that some of the servers have changed name and consequently the url in which we relied to identify if the file was already in the tree area. Initially I've modified my scripts to make sure for example that if the server was aims3.llnl.gov the script will also check files in pcmdi3{7/9}.llnl.gov . Then I noticed that dkrz changed server name too and handling all these exceptions it's too messy.
Not to say that PCMDI is not using the proper ensemble version but directories named 1/2/3 etc instead. Since the use of elasticsearch has been momentarily suspended, I'm looking again at using the sqlite database to handle this.
So I'm starting today to adapt my python script to download the search for data, update information on database including new fields for md5 and sha256 checksum and finally download the data if it isn't there.
Clearly adding md5/sha256 info means that we need to have a row for each file and so probably I'll have to split the database in different "experiments" or try to use postgres or mysql. Any suggestion and or comment on this is very welcome.
Instructions on how to use the blog
- To comment within previous blog topics
- Navigate to the blog topic of your choice
- Enter your comments at the bottom of the page
To create a new Blog Topic:
- Click the 'Create' button in the Confluence header bar above
- Choose the 'Blog Post' option (the default Space should be this CMIP Community wiki)